Abstract
Recent advances in learning multi-modal representation have witnessed the success in biomedical domains. While established techniques enable handling multi-modal information, the challenges are posed when extended to various clinical modalities and practical modality-missing setting due to the inherent modality gaps. To tackle these, we propose an innovative Modality-prompted Heterogeneous Graph for Omni-modal Learning (GTP-4o), which embeds the numerous disparate clinical modalities into a unified representation, completes the deficient embedding of missing modality and reformulates the cross-modal learning with a graph-based aggregation. Specially, we establish a heterogeneous graph embedding to explicitly capture the diverse semantic properties on both the modality-specific features (nodes) and the cross-modal relations (edges). Then, we design a modality-prompted completion that enables completing the inadequate graph representation of missing modality through a graph prompting mechanism, which generates hallucination graphic topologies to steer the missing embedding towards the intact representation. Through the completed graph, we meticulously develop a knowledge-guided hierarchical cross-modal aggregation consisting of a global meta-path neighbouring to uncover the potential heterogeneous neighbors along the pathways driven by domain knowledge, and a local multi-relation aggregation module for the comprehensive cross-modal interaction across various heterogeneous relations. We assess the efficacy of our methodology on rigorous benchmarking experiments against prior state-of-the-arts. In a nutshell, GTP-4o presents an initial foray into the intriguing realm of embedding, relating and perceiving the heterogeneous patterns from various clinical modalities holistically via a graph theory.
Insight
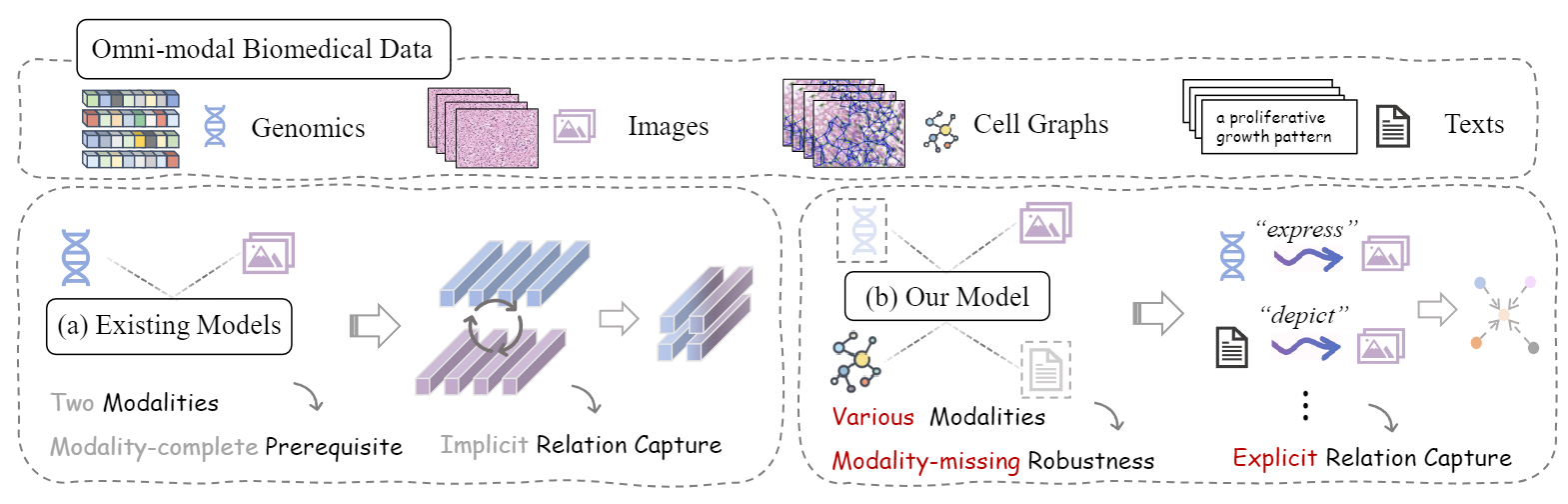
Overview: Unlike (a) prior methods, (b) our framework enables learning unified omni-modal representation from various clinical modalities with modality missing and explicit capture of the cross-modal relations through the established heterogeneous graph representation.
Framework
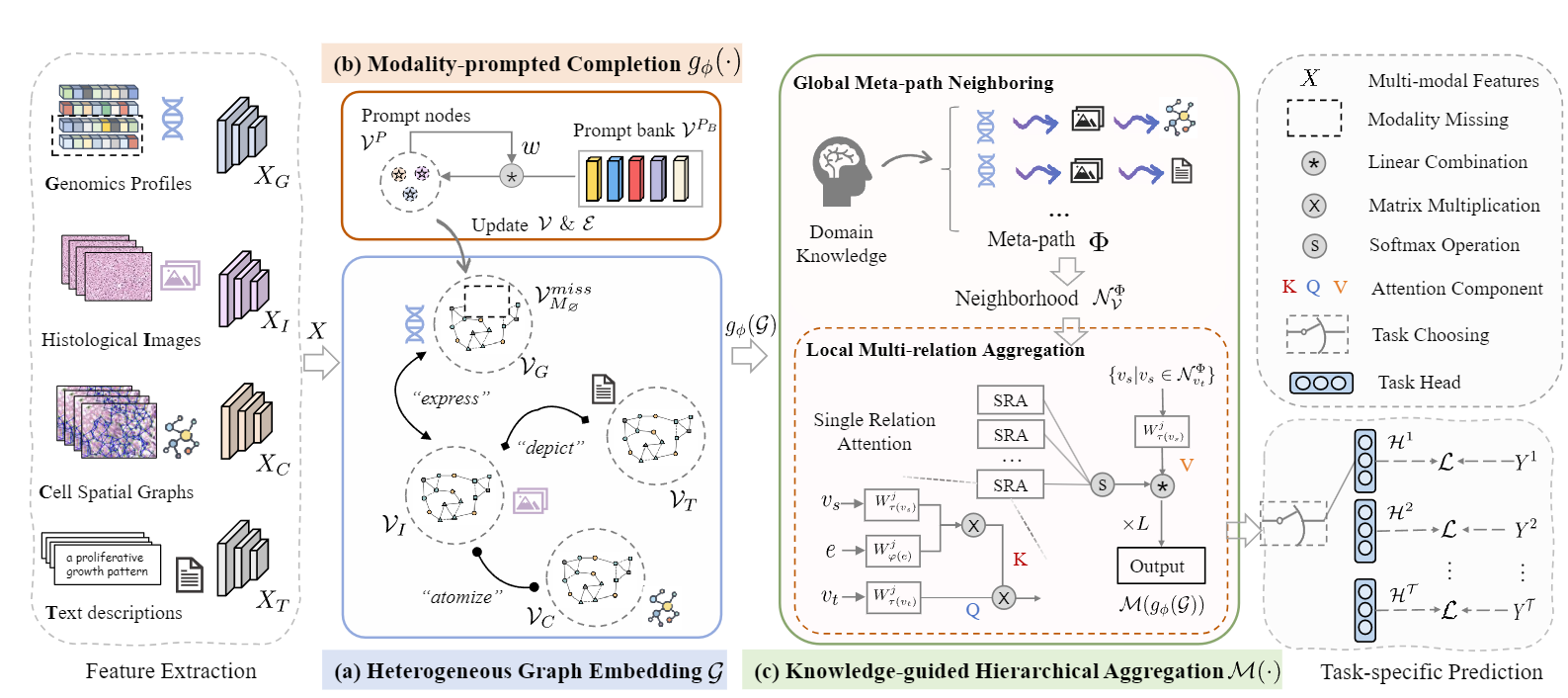
Pipeline: We instantiate the omni-modal biomedical features, and embed them onto (a) the heterogeneous graph space. Then, we introduce (b) the modality-prompted completion via graph prompting to complete the missing embedding. After that, we design (c) the knowledge- guided hierarchical aggregation from a global meta-neighbouring to uncover the het- erogeneous neighbourhoods and a local multi-relation aggregation to interact features across various heterogeneous relations.
Experiments
Performance Comparison
We report results on four TCGA benchmarks, using various modality combinations of Genomics, Images, Cell graphs and Texts.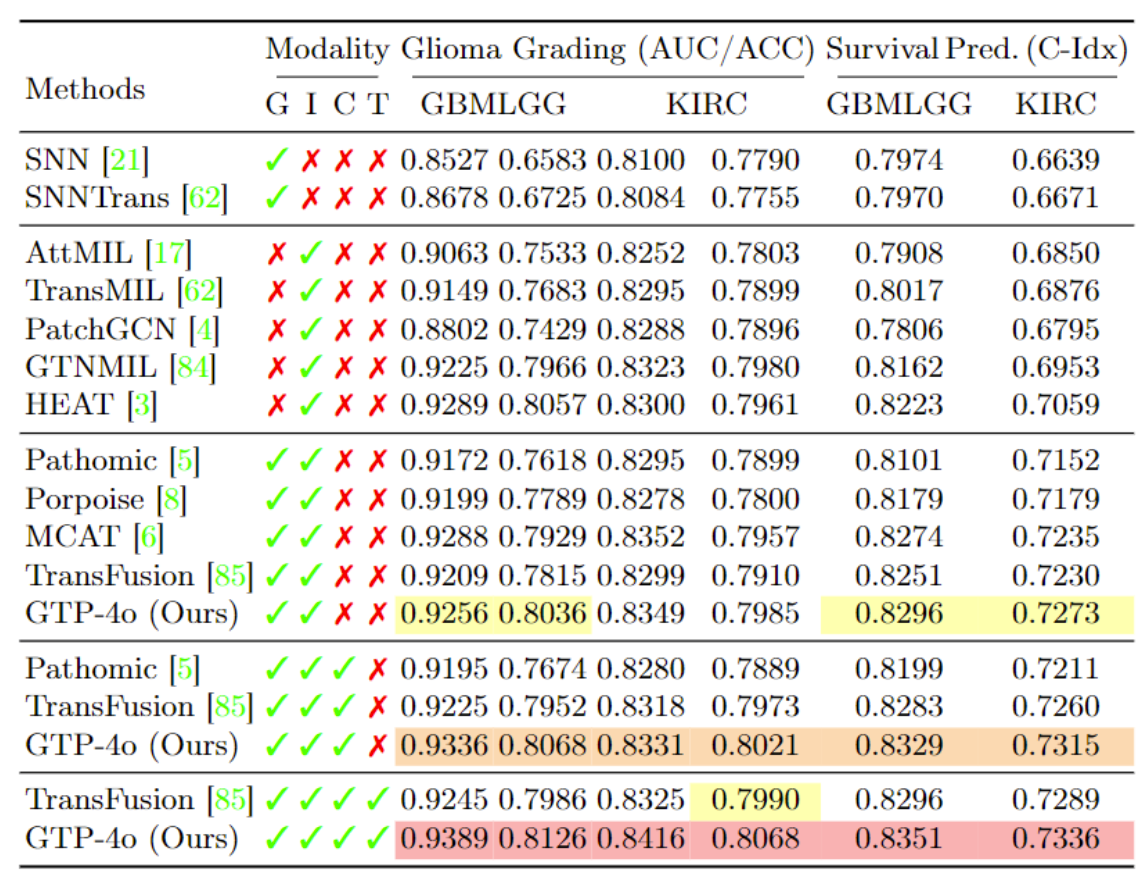
Ablation & Visualization
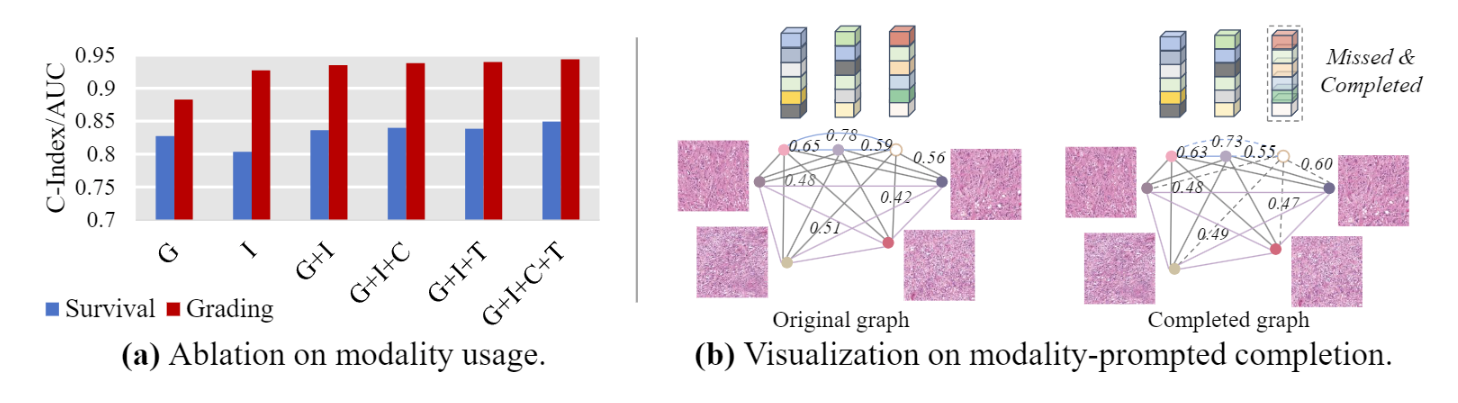 (a) Analysis of Modality Usage. We provide the results of GTP-4o by
using either Genes, Images, Cell graphs, Texts, or their combinations, on benchmarks of
survival prediction (C-Index) and glioma grading (AUC).
(a) Analysis of Modality Usage. We provide the results of GTP-4o by
using either Genes, Images, Cell graphs, Texts, or their combinations, on benchmarks of
survival prediction (C-Index) and glioma grading (AUC).
(b) Analysis of Modality- prompted Completion. We compare the relation pattern (similarity) in the original graph and the graph that is first removed specific instances then completed.